Automatic model selection for high-dimensional Cox models with MCP penalty, evaluated by penalized partial-likelihood.
Usage
fit_mcp(
x,
y,
nfolds = 5L,
gammas = c(1.01, 1.7, 3, 100),
eps = 1e-04,
max.iter = 10000L,
seed = 1001,
trace = FALSE,
parallel = FALSE
)Arguments
- x
Data matrix.
- y
Response matrix made by
Surv.- nfolds
Fold numbers of cross-validation.
- gammas
Gammas to tune in
cv.ncvsurv.- eps
Convergence threshhold.
- max.iter
Maximum number of iterations.
- seed
A random seed for cross-validation fold division.
- trace
Output the cross-validation parameter tuning progress or not. Default is
FALSE.- parallel
Logical. Enable parallel parameter tuning or not, default is
FALSE. To enable parallel tuning, load thedoParallelpackage and runregisterDoParallel()with the number of CPU cores before calling this function.
Examples
# \donttest{
data("smart")
x <- as.matrix(smart[, -c(1, 2)])
time <- smart$TEVENT
event <- smart$EVENT
y <- survival::Surv(time, event)
fit <- fit_mcp(x, y, nfolds = 3, gammas = c(2.1, 3), seed = 1001)
#> Warning: ncvsurv() is intended for pathwise optimization, not for single values of lambda.
#> 1. You are strongly encouraged to fit a path and extract the solution at the lambda value of interest, rather than use ncvsurv() in this way.
#> 2. In particular, if you are using the MCP or SCAD penalties, be aware that you greatly increase your risk of converging to an inferior local maximum if you do not fit an entire path.
#> 3. You may wish to look at the ncvfit() function, which is intended for non-path (i.e., single-lambda) optimization and allows the user to supply initial values.
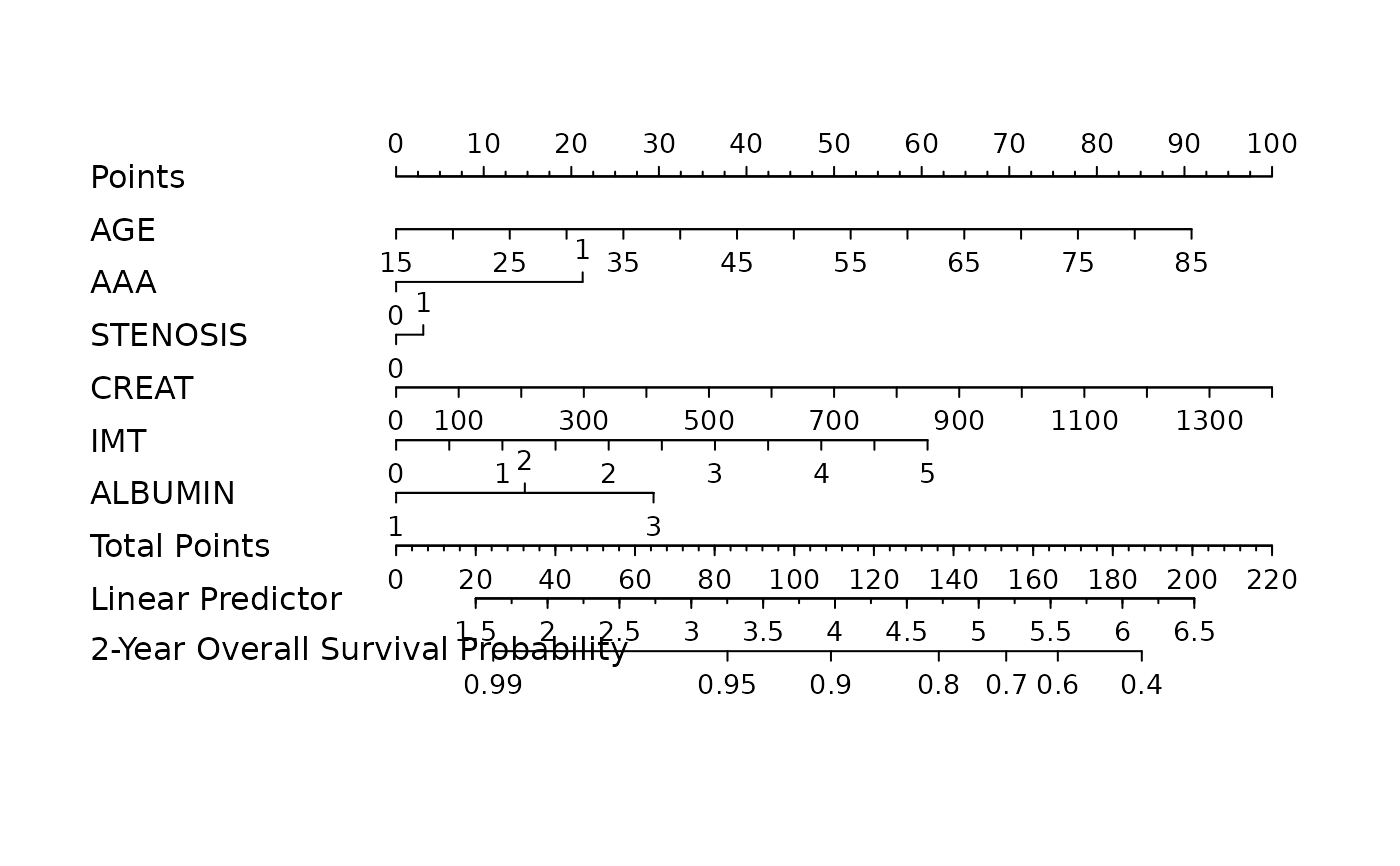
nom <- as_nomogram(
fit, x, time, event,
pred.at = 365 * 2,
funlabel = "2-Year Overall Survival Probability"
)
plot(nom)
 # }
# }
