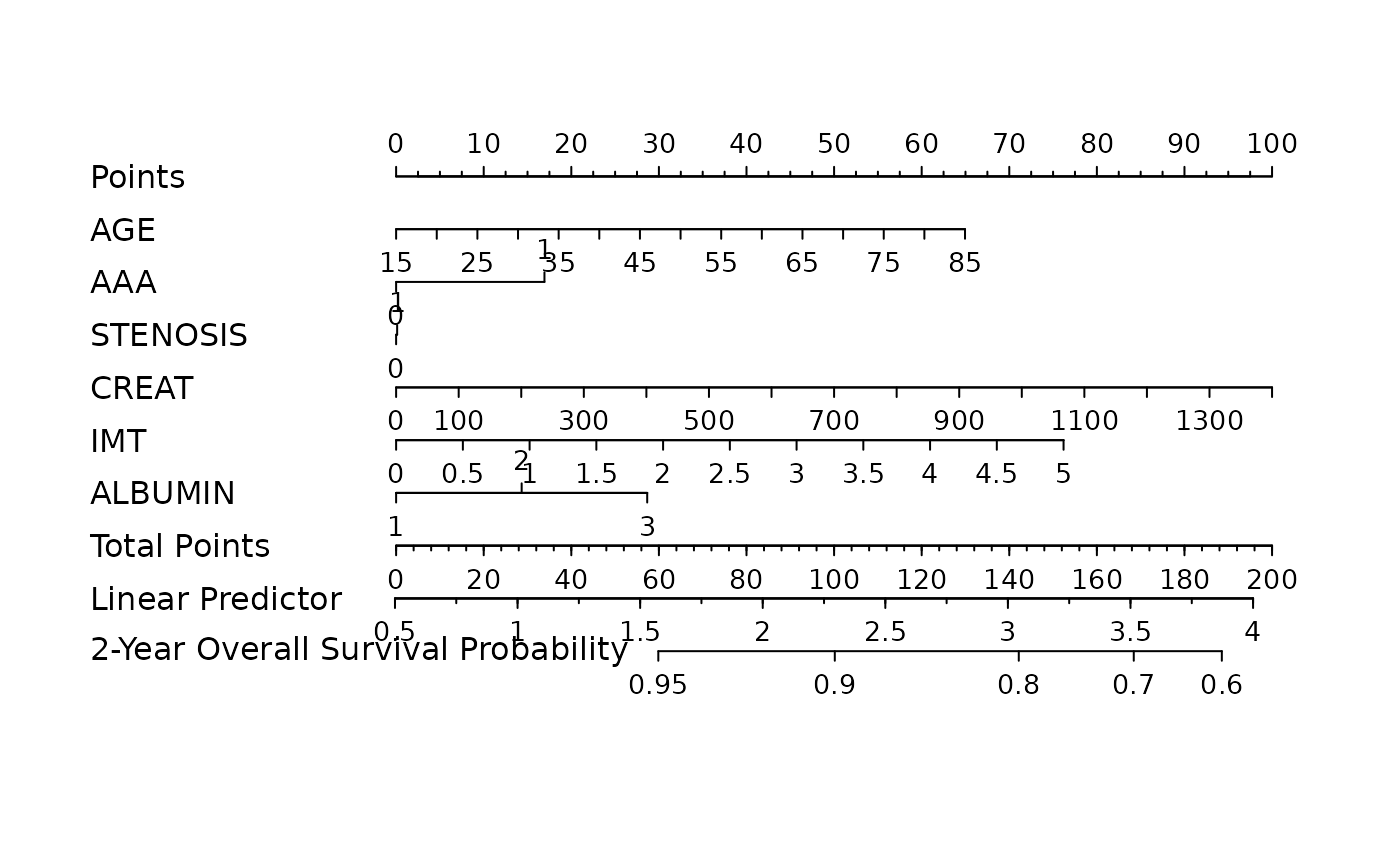
Construct nomograms ojects for high-dimensional Cox models
Arguments
- object
Model object fitted by `hdnom::fit_*()` functions.
- x
Matrix of training data used for fitting the model.
- time
Survival time. Must be of the same length with the number of rows as
x.- event
Status indicator, normally 0 = alive, 1 = dead. Must be of the same length with the number of rows as
x.- pred.at
Time point at which to plot nomogram prediction axis.
- fun.at
Function values to label on axis.
- funlabel
Label for
funaxis.
Note
The nomogram visualizes the model under the automatically selected "optimal" hyperparameters (e.g. lambda, alpha, gamma).
Examples
data(smart)
x <- as.matrix(smart[, -c(1, 2)])
time <- smart$TEVENT
event <- smart$EVENT
y <- survival::Surv(time, event)
fit <- fit_lasso(x, y, nfolds = 5, rule = "lambda.1se", seed = 1001)
nom <- as_nomogram(
fit, x, time, event, pred.at = 365 * 2,
funlabel = "2-Year Overall Survival Probability"
)
print(nom)
#> Points per unit of linear predictor: 100.9146
#> Linear predictor units per point : 0.009909369
#>
#>
#> AGE Points
#> 15 0
#> 20 5
#> 25 11
#> 30 16
#> 35 21
#> 40 27
#> 45 32
#> 50 38
#> 55 43
#> 60 48
#> 65 54
#> 70 59
#> 75 64
#> 80 70
#> 85 75
#>
#>
#> AAA Points
#> 0 0
#> 1 13
#>
#>
#> CREAT Points
#> 0 0
#> 100 7
#> 200 14
#> 300 21
#> 400 29
#> 500 36
#> 600 43
#> 700 50
#> 800 57
#> 900 64
#> 1000 71
#> 1100 79
#> 1200 86
#> 1300 93
#> 1400 100
#>
#>
#> IMT Points
#> 0.0 0
#> 0.5 4
#> 1.0 9
#> 1.5 13
#> 2.0 17
#> 2.5 22
#> 3.0 26
#> 3.5 30
#> 4.0 35
#> 4.5 39
#> 5.0 44
#>
#>
#> ALBUMIN Points
#> 1 0
#> 2 18
#> 3 36
#>
#>
#> Total Points 2-Year Overall Survival Probability
#> 122 0.90
#> 50 0.95
#>
plot(nom)