Model selection for high-dimensional Cox models with adaptive elastic-net penalty
Source:R/1_1_model.R
fit_aenet.RdAutomatic model selection for high-dimensional Cox models with adaptive elastic-net penalty, evaluated by penalized partial-likelihood.
Arguments
- x
Data matrix.
- y
Response matrix made with
Surv.- nfolds
Fold numbers of cross-validation.
- alphas
Alphas to tune in
cv.glmnet.- rule
Model selection criterion,
"lambda.min"or"lambda.1se". Seecv.glmnetfor details.- seed
Two random seeds for cross-validation fold division in two estimation steps.
- parallel
Logical. Enable parallel parameter tuning or not, default is
FALSE. To enable parallel tuning, load thedoParallelpackage and runregisterDoParallel()with the number of CPU cores before calling this function.
Examples
data("smart")
x <- as.matrix(smart[, -c(1, 2)])
time <- smart$TEVENT
event <- smart$EVENT
y <- survival::Surv(time, event)
# To enable parallel parameter tuning, first run:
# library("doParallel")
# registerDoParallel(detectCores())
# then set fit_aenet(..., parallel = TRUE).
fit <- fit_aenet(
x, y,
nfolds = 3, alphas = c(0.3, 0.7),
rule = "lambda.min", seed = c(3, 7)
)
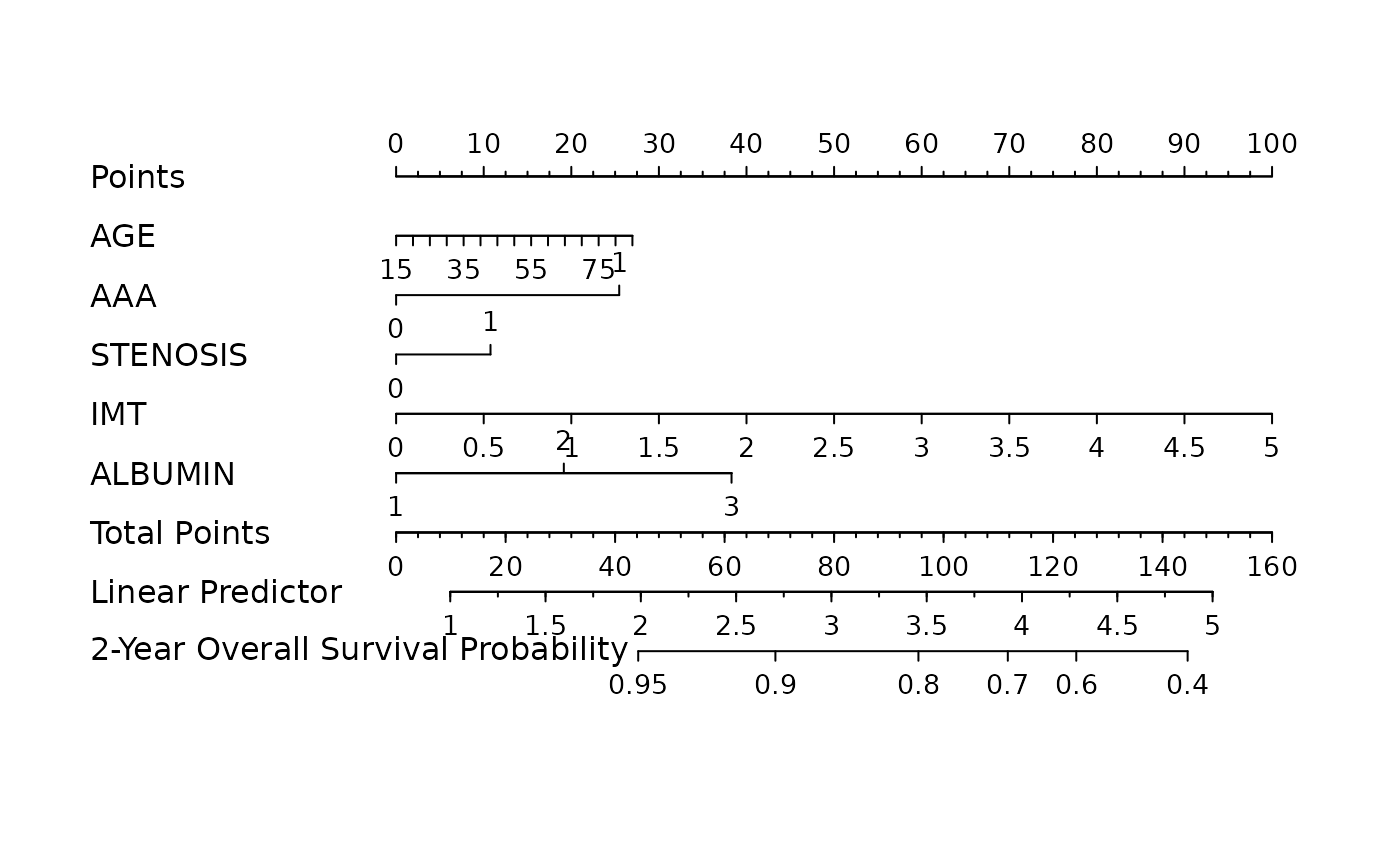
nom <- as_nomogram(
fit, x, time, event,
pred.at = 365 * 2,
funlabel = "2-Year Overall Survival Probability"
)
plot(nom)